Abstract
Lenalidomide is an immunomodulatory therapeutic (IMiD) with known anti-proliferative and anti-angiogenic effects, and is an approved therapy for both 5q- Myelodysplastic Syndromes and Multiple Myeloma (MM). This thalidomide analog is known to retarget the E3 ubiquitin ligase cereblon to differential ubiquitination, and therefore degradation, of specific substrates. While lenalidomide maintenance is known to promote progression-free survival, increased rates of therapy-related myeloid neoplasms (TMNs) have been observed in MM patients on IMiD maintenance therapy following autologous stem cell transplant. Of note, TP53 is the most commonly mutated gene in TMN arising after lenalidomide therapy (Mouhieddine et al. 2020). TP53 is also recurrently mutated in TMNs following chemotherapy, suggesting that genotoxic stress results in the selective expansion of hematopoietic stem and progenitor cells (HSPCs) carrying mutations in TP53 (TN Wong et al. 2018). Based on these observations, we hypothesize that lenalidomide may contribute to the development of TMN by providing a fitness advantage to HSPCs carrying TP53 mutations.
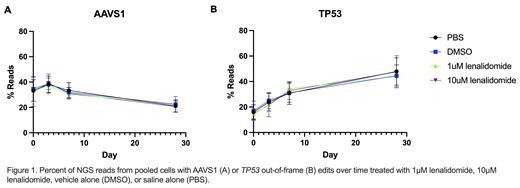
To test this hypothesis, we generated TP53 -/- deficient MOLM13 AML cells using CRISPR-Cas9 gene editing. We confirmed guide editing efficiency by next generation sequencing of a bulk edited pool of cells and generated isogenic clones via outgrowth of singly sorted cells, in an accepted approach for the field (Boettcher et al. 2019). We validated clones via sequencing and western blotting, and identified no differences in basal proliferation characteristics. Next, we further compared TP53 -/- deficient clones' with TP53 +/+ clones' response to cisplatin treatment (0.05-10µM) and observed a 2.8 fold increase in IC50 for TP53 -/- deficient clones. Satisfied that TP53 -/- isogenic clones successfully recapitulated this known resistance phenotype, we examined the response to lenalidomide by mixing TP53 -/- and TP53 +/+(AAVS1 edited control) clones and measuring expansion under lenalidomide treatment. However, we observed substantial inter-clone heterogeneity, with widely varying cellular proliferation characteristics following clone mixing, despite our prior validation. Therefore, we shifted to using bulk edited MOLM-13 cells in order to mitigate the potential impact of off-target effects and strong selective pressure of singly sorted clones. We edited a pool of cells at either TP53 or the safe harbor locus AAVS1 , allowed cells to recover, and mixed the two edited populations before treating with PBS, vehicle (DMSO), 1µm or 10µM lenalidomide for up to 28 days. We sequenced both AAVS1 and TP53 loci to measure cellular expansion by tracking the fraction of unedited, safe harbor edited, and TP53-/- genomic DNA at 0, 3, 7, and 28 days. Contrary to our hypothesis, TP53 -/- cells did not expand over time in response to lenalidomide treatment, and we did not detect a difference in TP53 -/- cell expansion with lenalidomide treatment compared to vehicle control (Figure 1). While we did not detect a cell intrinsic effect of TP53 mutations on lenalidomide response, it is possible that there are cell extrinsic effects, such as alterations in the bone marrow microenvironment, that contribute to the development of TP53-mutant TMN. It is also possible that a more prolonged exposure of lenalidomide is needed to see a significant effect of TP53-mutant cell expansion. This is relevant, since patients may be maintained on lenalidomide therapy for years.
No relevant conflicts of interest to declare.